Objective
and Overview
The objective of
this tutorial is to introduce users to
the GBSW implicit solvent model in CHARMM. GBSW stands for a
generalized Born (GB) model with a simple smoothed SWitching function.
In this
tutorial we will:
- Generate the
pentameric phospholamban by reading PDB:1ZLL into CHARMM, and orient
its transmembrane (TM) domain along Z,
- Solvate the
protein with GBSW implicit solvent
- Minimize,
equilibrate, and simulate the system with and without membrane
- use 5-fold
symmetry in the membrane simulation
Phospholamban is a transmembrane (TM) protein which regulates Ca2+
ATPase. The number of simulation steps is limited in this tutorial due
to the time constraint, but can be extended to explore the stability
and dynamics of the protein in solution as well as in membrane
environment.
Please
note that CHARMM supports various implicit membrane models such as
IMM1, GB/IM, and GBMV. (Note, to run this tutorial you need to download and unpack the pdb subdirectory and the parameters. You can also download the output files if you wish to check yourself.)
|
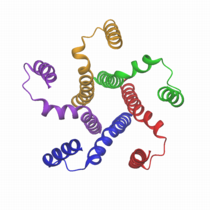
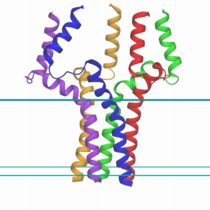
A molecular graphical
view of pentameric phospholamban in GBSW membrane implicit solvent
model.
|
GBSW simulations with phospholamban
(PDB:1ZLL)
| gbswmemb1.inp |
This
input illustrates a typical setup for simulations in GBSW membrane
without images. In this example, we will
- Read
the
coordinates of each monomer by generating their PSF in CHARMM.
- Orient the
TM domains along Z and adjust their position to place in the
hydrophobic core
coor
stat
coor
orient
coor
stat
coor
rotate ydir 1.0 phi 90.0
coor
trans zdir -?zmin
coor
trans zdir -17.0
- Read GBSW
optimal radii
prnlev
0
stream
radii_prot_na.str
stream
radius_gbsw.str
prnlev
5 node 0
define
check select (.not type H* ) .and. -
(
property wmain .eq. 0.0 )
show end
if
?nsel ne 0 stop !some heavy atom have a zero radius
- Setup GBSW
gbsw
sgamma 0.005 nang 50 dgp 1.5 tmemb
30.0 msw 2.5
where sgamma
is the surface
tension coefficient, nang
is
the number of
angular integration points, dgp
is a
grid spacing for lookup table, msw
is the half of switching
length over which hydrophobic environment
is changed to solvent region, and tmemb-msw*2
is the hydrophobic length
of the membrane, i.e., here it is 25 A.
- Minimize,
equilibrate, and let it go for sampling. One can certainly extend the
simulation by simply changing the number of simulation steps or
restarting it. Once the calculation is done, one can see the dynamics
of the protein using "vmd".
|
| gbswmemb2.inp |
This
input illustrates a typical setup for simulations in GBSW membrane
with images. In this example, we will use 5-fold symmetry along Z. Once
we read and orient all the five monomers as illustrated in
"gbswmemb1.inp", we will delete all the monomer except the
first
one;
delete
atom sele
.not. segid PROA end
The 5-fold symmetry is setup;
READ
IMAGE
*
5-fold symmetry
*
IMAGE
RT2
ROTATE
0.0 0.0
1.0 72.0
IMAGE
RT3
ROTATE
0.0 0.0 1.0
144.0
IMAGE
RT4
ROTATE
0.0 0.0 1.0
216.0
IMAGE
RT5
ROTATE
0.0 0.0 1.0
288.0
END
So, we only simulate one monomer, but there are interactions with
others (images) under the constraint of the symmetry. One has to put
the "image cutoff" in the nonbond specification;
!setup
nonbond
option for GBSW
calc
ctonnb = 16.0
calc
ctofnb =
@ctonnb
calc
cutnb =
@ctofnb + 4.0
calc
cutim =
@cutnb
NBOND
atom switch
cdie vdw vswitch -
ctonnb @ctonnb ctofnb @ctofnb cutnb @cutnb cutim @cutim
Once this is done, the rest is the same as gbswmemb1.inp. In other
words, once the images are setup, GBSW module automatically consider
the influence of the images.
One can setup any
kinds of images (in solvent and in membrane) before GBSW setup to
consider the influence of those images in GBSW calculations.
|
| gbsw.inp |
This
input illustrates a typical setup for simulations in GBSW solvent
without images. The input itself is identical to "gbswmemb1.inp" except
the GBSW setup without membrane option;
gbsw
sgamma 0.005 nang 50 dgp 1.5
|
written
by Wonpil Im
|
|



