These examples represent somewhat routine analyses. These
facilities, however, can be combined with the CHARMM scripting
language to achieve many less-"routine" analyses. While you play
with the examples, please examine the structure and the trajectory
using VMD. Also use your favorite graphing software to make diagrams
from the data files produced. All data plots shown in this page are
screen shots generated by the gnuplot scripts provided. Please also
keep in mind that the provided 10 ps trajectory with 100 coordinate
frames sampled every 50 fs is far too short for many of the
phenomena that we are going to look at.
To load the gnuplot script, first type "gnuplot" to start gnuplot.
Then at gnuplot prompt, type:
1. Recentering/orienting and removing water from the trajectory
orient.inp
The raw trajectory often needs to be pre-process to facilitate some
of the later analyses. The trajectrory was obtained from a
simulation using periodic boundary conditions (PBC), which means
that it is possible for the solute (the protein) to be partly out of
the primary simulation box if we look at the raw trajectory. By
recentering the trajectory we move solvent molecules, according to
the PBC, so that the protein is in the center of the box in each
frame. Furthermore we then re-orient each frame so that the protein
is superimposed on the coordinates of the initial protein structure,
thus removing overall protein rotation/translation motions. For a
very flexible system the removal of overall rotation may be a
non-trivial task, but grx1 is compact and quite rigid so we base the
superposition on all atoms in the protein.
The script uses the CHARMM MERGE command (dynamc.doc) and
demonstates only the part that we re-orient the protein and remove
all water molecules. A new trajectory file is produced. The script
also contains an example on how to re-center and orient the protein
without deleting water molecules (this line is not excuted as it is
after the "stop" command). MERGE can also be used to join or
split trajectory files or to change (reduce) sampling frequency.
Back to Top
2. Root Mean Square Deviation and Radius of Gyration
rmsd-singlepdb.inp
rmsd-rgyr-traj-correl.inp
rmsd-rgyr-traj-corman.inp
Two simple characteristics of a structure are i) its
RMSD wrt to some reference structure (eg, the
starting point of a simulation), which tells us something about the
dissimilarity between the structures - 1 Å RMSD is barely visible to
the eye, and RMSD 10 Å is very different, and ii) its RGYR,
which is a combined measure of its overall size and shape. A sphere
has the smallest RGYR of all bodies of the same volume increasing
the volume, making the shape more anisotropic or having more of its
mass at the periphery all lead to an increased RGYR. The
proper definition of RGYR in mechanics is mass-weighted, which you
can get by adding keyword MASS to the COOR RGYR command.
Three scripts are provided to demonstrated how one evaluate RMSD
and RGYR using CHARMM. Script "rmsd-singlepdb.inp" is extremely
simple and only calculates CA RMSD between two structures. Scripts
"rmsd-rgyr-traj-correl.inp" and "rmsd-rgyr-traj-corman.inp"
calculate RMSD and RGYR of a trajectory from a reference structure
using two methods (CORREL vs. loop with CORMAN). In CORREL, the
results could have been output to one file for each property, but we
use the ability to edit the dimensions of the extracted time series
in CORREL (edit ... veccod) to put all the information into one
file.
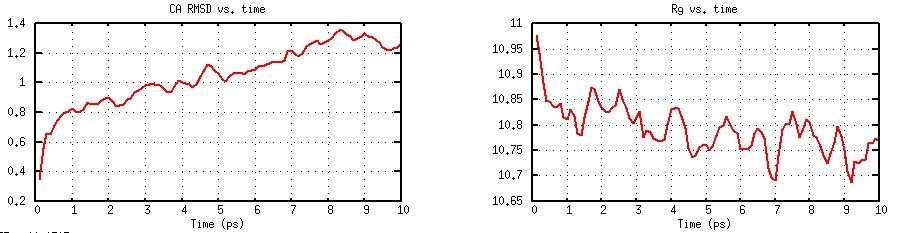
CA RMSD and Rg as functions of time (generated by
"plot_rmsd.gnu").
Back to Top
3. Hydrogen Bond analysis
hbond.inp
Hydrogen bonding patterns often provide useful information. In this
exercise we use the COOR HBOND (corman.doc) command to find hydrogen
bonds in a single structure, as well as average number of hydrogen
bonds and their average life times from a trajectory. Hydrogen bonds
can be defined in many ways, but we use a simple geometric
criterion: A hydrogen bond exists if the distance between the
hydrogen and acceptor atoms is less than 2.4 Å. This works well in
practice. Note that when we have information about the hydrogen
position, the hydrogen bond definition is not very sensitive to
angular criteria, as are often used when determining hydrogen bonds
in X-ray structures (lacking hydrogens). We will look at hydrogen
bonds within the protein, between protein and water, and also on
water-mediated hydrogen bonding contacts between different parts of
the protein, ie hydrogen bonded interactions of the form A/D
- water - A/D, where A/D denotes a hydrogen bond donor or acceptor
in the protein. COOR HBOND uses the information about acceptors and
donors in the PSF so we can use quite simple, and general
atom-selections; all hydrogen bonds between the acceptors/donors in
the two selections are calculated. A second form of the command
(COOR CONTact) just applies the distance criterion to all the
selected atoms.
The results are presented as hydrogen bonds per each acceptor/donor
in the first atom-selection; if the VERBose keyword is present a
break-down in terms of accptors/donors in the second atom-selection
is given. NB! The VERBose keyword is not useful in a CHARMM
loop when you want to extract total number of hydrogen bonds through
CHARMM variables (subst.doc). <occupancy> is the average
number of hydrogen bonds formed by a given accptor/donor during the
trajectory. The <lifetime> (in ps) is the average
duration of each instance of a given hydrogen bond.
Back to Top
4. Secondary Structure
2nd-structure.inp
COOR SECS (corman.doc) computes the secondary structure
characteristics of a protein using the DSSP algorithm (Kabsch and
Sander 1983), which is based on backbone hydrogen bond patterns. The
CHARMM implementation is a slight simplification and uses the
hydrogen-acceptor-atom definition of a hydrogen bond. Results are
summarized in the output file (e.g., see below), and are also
returned as a numerical flag in the WMAIN array. See
"plot_nmr-s2.gnu" for an example where we plot WMAIN together with
the order parameters from NMR relaxation analysis. The screen shot
is show in NMR section below.
...
CHARMM> COOR SECS SELE .not. resn tip3 end VERBOSE
SELRPN> 855 atoms have been selected out of 17088
Secondary structure (Kabsch&Sander) analysis.
Using 56 aa in a context of 56 aa.
14 aa in alpha-helix ( 25%), and 24 aa in beta-strands ( 42%).
1: MET-THR-TYR-LYS-LEU-ILE-LEU-ASN-GLY-LYS-THR-LEU-LYS-GLY-GLU
E E E E E E E E E E
16: THR-THR-THR-GLU-ALA-VAL-ASP-ALA-ALA-THR-ALA-GLU-LYS-VAL-PHE
E E E E H H H H H H H H
31: LYS-GLN-TYR-ALA-ASN-ASP-ASN-GLY-VAL-ASP-GLY-GLU-TRP-THR-TYR
H H H H H H E E E E
46: ASP-ASP-ALA-THR-LYS-THR-PHE-THR-VAL-THR-GLU-
E E E E E E
...
Back to Top
5. Phi/Psi Distributions
phi-psi-dist.inp
Peptide backbone dihdrals phi and psi determine the overall fold of
a protein. In this example we extract histograms of phi and psi
distribution from the trajectory for any selected residue using
correl.doc. This script has the ability to take commandline
arguments. The argument @rid has a
default value of 41 by the virtue of the following command in the
"phi-psi-dist.inp":
if @?rid eq 0 set rid = 41
However, the value of @rid can be
overwritten by following CHARMM excution:
$CHARMMEXEC rid=15 < phi-psi-dist.inp | tee phipsi.out
After the TRAJ command in the CORREL module has been executed
averages and fluctuations are printed out for each time
series. Both histograms are output to a single file named
"phi-psi-dist.dat", and the phi/psi time series are output to
"phipsi.dat". Following is a plot of of the results for Gly41.
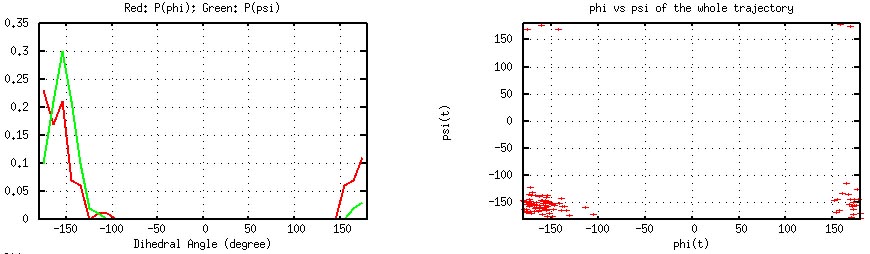
Gly41 phi/psi histograms (left) and time series (generated by
"plot_phipsi.gnu").
Back to Top
7. NMR Relaxation Rates and Generalized Order Parameters
nmr.inp
MD simulations and NMR relaxation experiments often cover similar
time-scales (ps-ns). Relaxation phenomena observed in NMR
experiments depend on motional behavior of nuclear dipoles in the
macromolecule, and the NMR module in CHARMM has been designed to
allow efficient extraction of NMR related parameters from a
trajectory (nmr.doc). This example analyzes relaxtaion parameters
(relaxation rates, generalized order parameters, configurational
entropy estimates) for the backbone amide groups.
Results, with some details such as the computed correlation
functions are given in the output file "nmr-rt_ct.dat", and are also
summarized in "nmr-r1r2.dat". Note that we use the trajectory with
overall protein (translation)/rotation removed - we assume that
internal and overall motions occur on very different time scales so
that we can confidently separate these motions and focus on the
internal motions. Such an assumption is necessary as most
simulations are not long enough to sufficient sample global
rotaional diffusion. The following figure plots the general order
parameter S2 for each HN vector together with the secondary
structure (generated by "2nd-structure.inp", see above). Clearly,
even for a 10 ps simulation, it is evident that loops are more
flexible (with smaller S2) (what a surprise. :D)
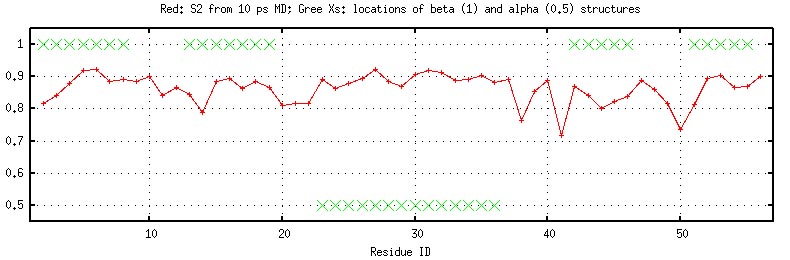
General order parameter and secondary structures
(generated by "plot_nmr-s2.gnu").
Back to Top
9. Phi/Psi Based Clustering
cluster.inp
As shown above in phi/psi distribution analysis, CORREL
(correl.doc) can be used to extract time series of backbone dihedral
angles (and many other properties). We can then use some of the
dihdrals that vary during the simulation for clustering the
structures in the trajectory. As shown from the rmsd analysis, the
protein does not move much with 10 ps. We thus choose to use loop
38-41 that has the largest fluctation (or smallers order parameters
from nmr analysis) to demonstrate clustering. The number of clusters
and cluster centers are summarized in "clus-center.dat". Membership
and distance from center of each frame are output in
"clus-member.dat".
Back to Top
Further Examples
The above exercises have been meant to show some of the analysis capabilities
of CHARMM. Most important is that you have to formulate your own scientifically
relevant questions, and then find a way to answer them. CHARMM is an evolving
research tool, and by combining its existing analysis facilites in various ways
using the scripting language you get a long way towards obtaining the data that
you need. More examples can be found at the CHARMM Web-site
www.charmm.org, in the Script Archive forum,
to which everybody is encouraged to submit their own scripts, for analysis,
structure manipulation, simulation or ....



