|
Objective and Overview
The objective of this tutorial is to
learn how to use the MMTSB Tool Set for carrying out simulations with NAMD.
NAMD is especially well-suited for
carrying out long simulations of large systems in explicit solvent because of good scaling on parallel architectures.
NAMD's input and output is compatible with CHARMM and it is therefore possible to first setup and equilibrate simulations
with CHARMM, carry out the production run with NAMD, and use CHARMM again for trajectory analysis.
The MMTSB Tool Set offers mdNAMD.pl as an interface to NAMD similar to the CHARMM interface mdCHARMM.pl. Typical usage of
mdNAMD.pl is introduced in this tutorial.
|
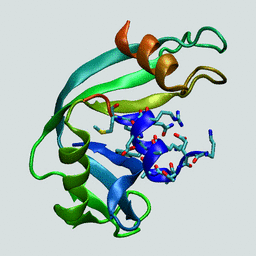
In this tutorial the N-terminal peptide from RNase (C-peptide) is used again. (see corresponding CHARMM tutorial here). An experimental structure is available as 1RNU from the Protein Data Bank. The structure is shown on the right with the C-peptide shown with sidechains.
|
|
1. Initial Structure
Begin by downloading/copying the PDB file for the structure of RNase (1RNU). Extract the C-peptide residues (1 through 13)
from the PDB file with:
convpdb.pl -sel 1:13 1RNU.pdb > 1rnu_cpep.pdb
and complete missing hydrogens by using complete.pl:
complete.pl 1rnu_cpep.pdb > 1rnu_cpep.allh.pdb
Check the result and verify that the structure is complete and looks like a helix.
2. Energy Minimization in Vacuum
Next, the initial strucute is briefly minimized in vacuum using CHARMM with restraints on the C-alpha coordinates (with a force constant
of 1 kcal/mol/Å2):
minCHARMM.pl -par minsteps=50 -cons ca self 1:13_1.0 -log min.log 1rnu_cpep.allh.pdb > 1rnu_cpep.min.pdb
This command will initially carry out steepest descent minimization and then switch to the more effective
adopted-basis Newton-Raphson minimization method over 50 steps. The resulting minimized conformation is redirected into a new file
(1rnu_cpep.min.pdb). CHARMM output during the minimization is available in min.log
3. Solvation
Next, the minimized structure is solvated in a cubic box of water. This is accomplished with the following command:
convpdb.pl -solvate -cubic -cutoff 9 1rnu_cpep.min.pdb > 1rnu_cpep.solvated.pdb
The cutoff determines the thickness of the water layer from the peptide to the edge of the box.
Note the box size that was written on the screen during the last command. It should be near 41 Å. We will need it as
input later when starting the NAMD simulation.
The peptide has a net charge of +1. In order to neutralize the system we have to add one Cl- counterion. This is done by randomly
replacing one of the water molecules with the following command:
convpdb.pl -ions CLA:1 1rnu_cpep.solvated.pdb > 1rnu_cpep.solvions.pdb
The PDB file 1rnu_cpep.solvions.pdb contains the peptide, water molecules, and a single chlorine ion. This file could be used for
simulations with CHARMM but needs to manipulated further for NAMD. NAMD expects water molecules at the end of the PDB file and has problems
with ions at the end of the PDB. We will use the following commands to first extract the peptide/water/ion parts into separate PDB files, each
with a different chain ID, and then reassemble them in the order peptide/ion/water so that NAMD can handle the file:
convpdb.pl -nsel peptide -setchain A 1rnu_cpep.solvions.pdb > solute.chainA.pdb
convpdb.pl -nsel CLA -setchain I 1rnu_cpep.solvions.pdb > ions.chainI.pdb
convpdb.pl -nsel water -setchain W 1rnu_cpep.solvions.pdb > water.chainW.pdb
convpdb.pl -merge ions.chainI.pdb solute.chainA.pdb | convpdb.pl -merge water.chainW.pdb > 1rnu_cpep.solvions.namd.pdb
4. Energy Minimization and Initial Equilibration with NAMD
The fully solvated system can now be minimized and heated up with NAMD.
NAMD requires two input files, a structure in PDB format and a CHARMM PSF file to describe the molecular
topology and force field parameters such as charges. A PSF file is generated from the equilibrated structure using CHARMM with the following command:
genPSF.pl -xplor 1rnu_cpep.solvions.namd.pdb > 1rnu_cpep.psf
Then the following command accomplishes minimization (for 50 steps) and an initial heating run at 50K with NAMD
(replace xxx with the boxsize):
mdNAMD.pl -par dynsteps=500,boxsize=xxx,dyntemp=50,minsteps=50 \
-restout equi.50 -log equi.50.out -cmd equi.50.inp 1rnu_cpep.psf 1rnu_cpep.solvions.namd.pdb
Once this command has finished you will find the NAMD input file in equi.50.inp, the NAMD output file
in equi.50.out and the coordinates, velocities, and box dimensions of the final conformation in
equi.50.coor, equi.50.vel, and equi.50.xsc, respectively. The default ensemble when
running with NAMD is NPT, so that the box size fluctuates during the simulation.
The equlibration is continued at a higher temperature with the following command:
mdNAMD.pl -par dynsteps=500,dyntemp=200 -coor equi.50.coor -ext equi.50.xsc \
-restout equi.200 -log equi.200.out 1rnu_cpep.psf 1rnu_cpep.solvions.namd.pdb
The initial structure is now read from equi.50.coor and the box information from
equi.50.xsc. A PDB file is still required at the end of the command, but the
coordinates are taken from the 'coor'-file instead.
If we want to continue at the same temperature (and keep the velocities) to run a short
production run we can do that with the following command:
mdNAMD.pl -par dynsteps=1000,dynoutfrq=50 -restart equi.200 \
-restout prod.1 -trajout prod.1.dcd -log prod.1.out 1rnu_cpep.psf 1rnu_cpep.solvions.namd.pdb
This command will read the coordinates, velocities, and box size information from the equi.200
run and continue for another 1000 steps. This command also illustrates how to write snapshots to
a trajectory file (in CHARMM DCD format) every 50 steps. The NAMD DCD file is fully compatible with
CHARMM and can be visualized in VMD and analyzed with CHARMM and other tools just like CHARMM trajectories.
5. Running in parallel with NAMD
A main reason for using NAMD instead of CHARMM is that it scales better for
large systems and on large parallel computers. NAMD can be run in parallel through mdNAMD.pl by simply changing a few environment
variables and setting up a host file with available CPUs.
We will begin with the host file. Use a text editor to generate a file that contains
group main
on the first line followed by
host name
for each CPU/core that is available either on the same machine or a remotely-accessible machine
where 'name' has to be replaced with the actual host name.
For example, if you are using a dual quad-core workstation with a total of 8
cores with the name snoopy, you would generate the following host file:
group main
host snoopy
host snoopy
host snoopy
host snoopy
host snoopy
host snoopy
host snoopy
host snoopy
Save this file under the name hostfile.
Now we set two environment variables. The first tells NAMD how to connect to
to other nodes:
export CONV_RSH=ssh
The previous command works on bash, if you are using csh or tcsh as your shell the command is:
setenv CONV_RSH ssh
The second environment variable describes how to run NAMD in parallel by changing
the NAMDEXEC variable. The following is a typical example:
export NAMDEXEC="/apps/namd/charmmrun ++nodelist hostfile +p8 /apps/namd/namd2 +giga"
In this case, NAMD is installed under /apps/namd. The path needs to be adjusted according to your setup. The host file that
we have created above is given with the ++nodelist option and in this case we want to run on 8 cores.
After setting both of these environment variables, we can now use mdCHARMM.pl to extend the previous simulation further with
the following command:
mdNAMD.pl -par dynsteps=5000,dynoutfrq=50 -restart prod.1 \
-restout prod.2 -trajout prod.2.dcd -log prod.2.out 1rnu_cpep.psf 1rnu_cpep.solvions.namd.pdb
Check the log file to see whether you are really running in parallel. For a small number of cores/CPUs, especially
on the same machine, you should see near-linear scaling, i.e. with 8 cores the simulation will only take 1/8th of
the time compared to a run on a single core.
|



